Just another bioinf blog
Blog by Michael Roach: @BeardyMcJohnFace@genomic.social
email: mroach@bioinf.cc
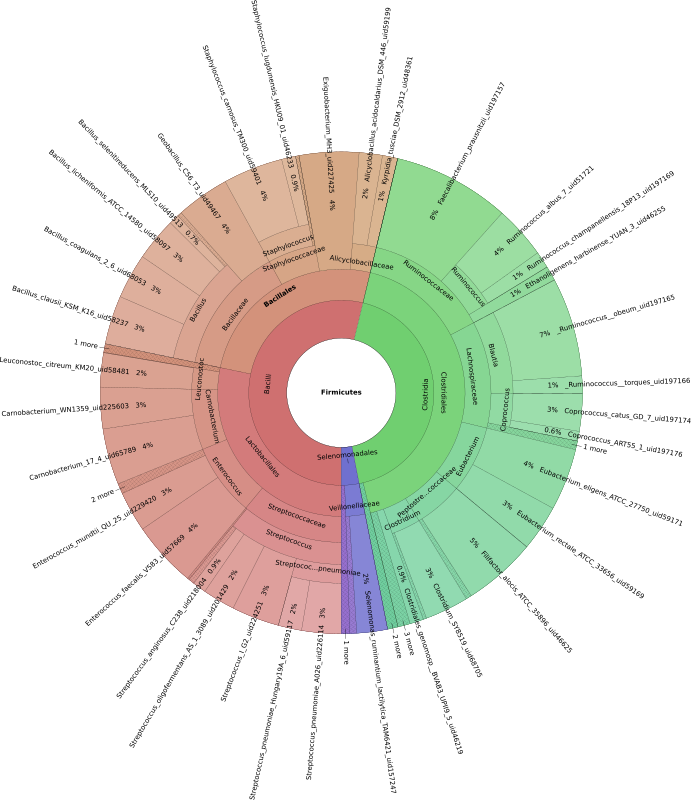
Krona plots are easy (metagenomics example)
21 Feb 2022
Krona plots are a fantastic way of representing heirarchial data such as taxonomic annotations. These plots are interactive, visually appealing, and best of all they're surprisingly easy to make. We'll use output from the metagenomics profiler 'FOCUS' as an example.FOCUS is an excellent tool for profiling the microbial communities of metagenomics samples, and its spiritual successor SuperFOCUS extends the tool to include subsystem-level profiling. This example will use a small output file: focusOutputEg.csv.zst
A quick look at this file shows columns 1-8 contain the taxonomic annotation, and columns 9 and 10 contain the percentage of R1 and R2 reads that match this annotation. BTW, if you’re not using zstandard, you should try it out. It has fast, parallel, decent file compression, and it has blazingly fast decompression–almost as fast as lz4.
$ zstdcat focusOutputEg.csv.zst | head
Kingdom,Phylum,Class,Order,Family,Genus,Species,Strain,ERR1467153_R1.fastq,ERR1467153_R2.fastq
Bacteria,Proteobacteria,Alphaproteobacteria,Rhodospirillales,Rhodospirillaceae,Rhodospirillum,Rhodospirillum_photometricum,Rhodospirillum_photometricum_uid159003,0.0,0.30543199636450163
Archaea,Euryarchaeota,Halobacteria,Halobacteriales,Halobacteriaceae,Natronobacterium,Natronobacterium_gregoryi,Natronobacterium_gregoryi_SP2_uid74439,0.12957821822784493,0.0805041736315925
Bacteria,Proteobacteria,Deltaproteobacteria,Desulfobacterales,Desulfobacteraceae,Desulfatibacillum,Desulfatibacillum_alkenivorans,Desulfatibacillum_alkenivorans_AK_01_uid58913,0.4362273642891679,0.2723663810922735
Bacteria,Proteobacteria,Betaproteobacteria,Burkholderiales,Comamonadaceae,Acidovorax,Acidovorax_sp._JS42,Acidovorax_JS42_uid58427,1.774572695525898,2.832211940191908
Bacteria,Proteobacteria,Alphaproteobacteria,Rhizobiales,Bradyrhizobiaceae,Rhodopseudomonas,Rhodopseudomonas_palustris,Rhodopseudomonas_palustris_BisA53_uid58445,0.0,0.16279122445280586
Bacteria,Firmicutes,Clostridia,Clostridiales,Peptostreptococcaceae,Clostridium,[Clostridium]_difficile,Clostridium_difficile_CF5_uid158359,0.05616259811855688,0.07011765537568315
Bacteria,Proteobacteria,Deltaproteobacteria,Myxococcales,Myxococcaceae,Corallococcus,Corallococcus_coralloides,Corallococcus_coralloides_DSM_2259_uid157997,0.0,0.03792781030247689
Bacteria,Proteobacteria,Gammaproteobacteria,Enterobacteriales,Enterobacteriaceae,Enterobacter,Enterobacter_sp._R4-368,Enterobacter_R4_368_uid208672,0.13149443908001496,0.0
Bacteria,Firmicutes,Clostridia,Clostridiales,Eubacteriaceae,Eubacterium,Eubacterium_eligens,Eubacterium_eligens_ATCC_27750_uid59171,0.5336824177251188,0.8448355470993689
For Krona, we want to use the text input file format. We will remove the header, have the count (sum of the R1 and R2 percent) for each line as the first column, and have tab-separated fields for the taxonomic annotations. We can do this with a bash command:
zstdcat focusOutputEg.csv.zst \
| tail -n+2 \
| awk -F ',' '{n=$9+$10; print n"\t"$1"\t"$2"\t"$3"\t"$4"\t"$5"\t"$6"\t"$7"\t"$8}' \
> focusKronaText.tsv
And look at our lovely new file:
$ head focusKronaText.tsv
0.305432 Bacteria Proteobacteria Alphaproteobacteria Rhodospirillales Rhodospirillaceae Rhodospirillum Rhodospirillum_photometricum Rhodospirillum_photometricum_uid159003
0.210082 Archaea Euryarchaeota Halobacteria Halobacteriales Halobacteriaceae Natronobacterium Natronobacterium_gregoryi Natronobacterium_gregoryi_SP2_uid74439
0.708594 Bacteria Proteobacteria Deltaproteobacteria Desulfobacterales Desulfobacteraceae Desulfatibacillum Desulfatibacillum_alkenivorans Desulfatibacillum_alkenivorans_AK_01_uid58913
4.60678 Bacteria Proteobacteria Betaproteobacteria Burkholderiales Comamonadaceae Acidovorax Acidovorax_sp._JS42 Acidovorax_JS42_uid58427
0.162791 Bacteria Proteobacteria Alphaproteobacteria Rhizobiales Bradyrhizobiaceae Rhodopseudomonas Rhodopseudomonas_palustris Rhodopseudomonas_palustris_BisA53_uid58445
0.12628 Bacteria Firmicutes Clostridia Clostridiales Peptostreptococcaceae Clostridium [Clostridium]_difficile Clostridium_difficile_CF5_uid158359
0.0379278 Bacteria Proteobacteria Deltaproteobacteria Myxococcales Myxococcaceae Corallococcus Corallococcus_coralloides Corallococcus_coralloides_DSM_2259_uid157997
0.131494 Bacteria Proteobacteria Gammaproteobacteria Enterobacteriales Enterobacteriaceae Enterobacter Enterobacter_sp._R4-368 Enterobacter_R4_368_uid208672
1.37852 Bacteria Firmicutes Clostridia Clostridiales Eubacteriaceae Eubacterium Eubacterium_eligens Eubacterium_eligens_ATCC_27750_uid59171
3.0158 Bacteria Proteobacteria Betaproteobacteria Methylophilales Methylophilaceae Methylotenera Methylotenera_versatilis Methylotenera_301_uid49469
Install KronaTools if you haven’t already using conda:
conda install -c bioconda krona
Run Krona. There are many KronaTools commands, but we’ll use ktImportText for converting out text input format into a Krona plot. The command is very simple:
ktImportText focusKronaText.tsv -o focusKrona.html
You can now open the .html file and explore your data. Double click to expand, back arrows to go back, download SVG snapshots for your publications and presentations!